“Commitment to the Quality”
We are dedicated to providing our customers with the highest-quality products available. We test our products from our network of trusted global suppliers to assure they meet our quality specifications. We stand behind all of our products knowing that our reputation lies on the success of your experiments.
Luciferin-Bioluminescence
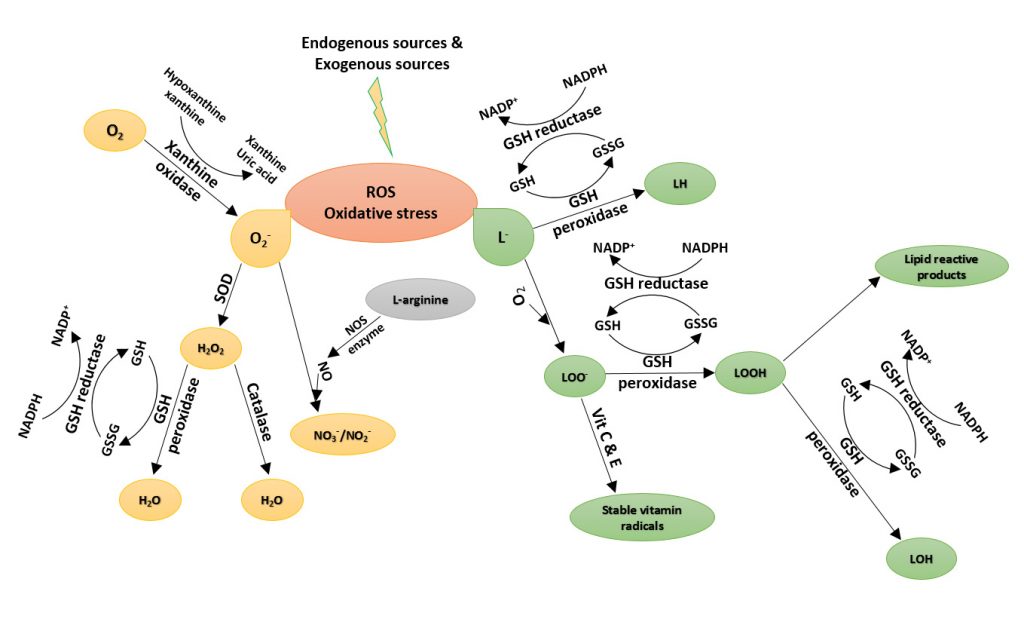
Bioluminescence is the production and emission of light by a living organism. Bioluminescence occurs widely in marine vertebrates and invertebrates, as well as in some fungi, microorganisms and terrestrial invertebrates. Luciferin, the firefly substrate of luciferase, is a common reagent used throughout the biotechnology field. The injection of luciferin allows for the real-time, noninvasive monitoring of disease progression and/or drug efficacy in these model systems through Bioluminescence Imaging (BLI). Coelenterazine is a luminophore is the substrate for many luciferase enzymes, including Renilla reniformis, gaussia, and aequorins. It is commonly used to monitor reporter genes in BRET (Bioluminescence Resonance Energy Transfer), ELISA and HTS methods. Gold Biotechnology’s luciferin and coelenterazine are Proven and Published™ to ensure a quality reagent with optimal potential.


IPTG
IPTG is an analog of galactose that is nonmetabolizable and inactivates the lac repressor to induce synthesis of β-galactosidase in E. coli. The expression of cloned genes under the control of the lac operon is induced by IPTG. IPTG is often used in the induction of recombinant proteins, is a substrate for thigalactoside transacetylase and has been reported to induce penicillinase in bacteria. IPTG is commonly used in cloning procedures that require induction of β-galactosidase and is most often used with X-Gal for blue/white bacterial colony screening. GoldBio’s is most cost effective IPTG on the market, without sacrificing quality or purity. Gold Biotechnology IPTG is Proven and Published™ and is a better way to generate the experimental results you need while focusing your budget on the things that really matter.
TCEP
TCEP (Tris (2-Carboxyethyl) phosphine Hydrochloride) is an odorless reducing agent suitable for protein applications and in the preparation for electrophoresis. TCEP is water soluble, is stable in a variety of solutions and is particularly useful in the labeling of cysteine residues with maleimides, preventing the cysteines from forming disulfide bonds. As a reducing agent, TCEP has many other desirable features, including: TCEP-HCl is stable in air. TCEP-HCl is odorless so reactions can be carried out on the bench without the need for a fume hood. TCEP-HCl is efficient and can be used to reduce disulfide bonds at low concentrations. TCEP-HCl may be used as a substitute for DTT at a final concentration of 50mM.

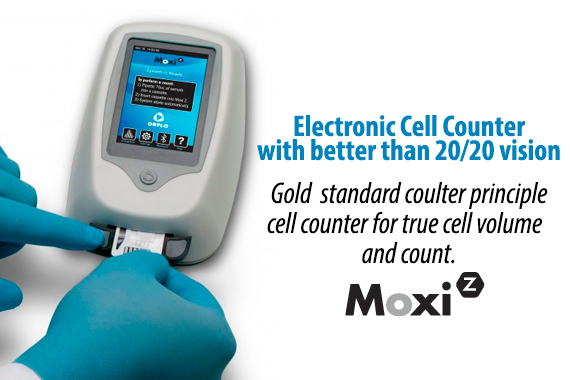
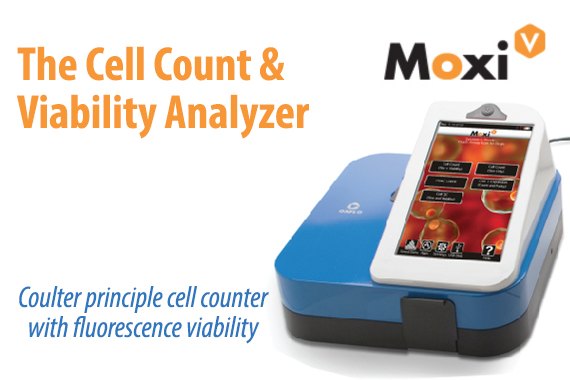
Higher Performance & Simplicity in One Disposable Cassette.
Patent-protected thin-film cassettes provide highly accurate, repeatable cell counts and cell size analysis in under 15 seconds. Inspired by the gold-standard Coulter Principle used in high-end counting systems worldwide, the Moxi cassette flows thousands of cells per test through a cell sensing zone to accurately capture the impedance-based volumetric measurement of each individual cell in the sample. Impedance-based cell counting of individual cells is proven to be over 95% accurate versus a 75% accuracy with image-based cell counting methods. No system contamination and integrated pre-filter for clogging prevention. Moxi cassettes are designed with a unique “”cell sieve”” to minimize cell clumping and clogging. In addition, Moxi cassettes contain 100% of your sample within the cassette body, eliminating the possibility for system contamination and sterilization. Cassettes are simply and easily discarded when testing is finished. Two tests per cassette. Each Moxi cassette offers two individual tests. The instrument will also alert the user if a used cassette is mistakenly inserted in the unit.
Healthcare, pharmaceutical and industrial organizations rely on quality-assured prepared media.
Maintaining cleanrooms and other aseptic areas to the most stringent regulations, plus validating sterility testing processes, requires quality assured prepared culture media. The EU GMP Annex 1 highlights an increasing need for organisations operating in pharmaceutical and medical device manufacture, and other industrial sectors, to reduce the risk of contamination wherever possible. Businesses that rely on clean room areas for their manufacturing will need to use prepared cultured media including poured media plates, bottled media, broth bags, ampoules – and perhaps even some bespoke media and accessories – to match the specific requirements of both organisation and facility.